AtaGenix Laboratories
AtaGenix Laboratories
Release time: 2025-10-29 View volume: 572
In October, AtaGenix continued to provide core technical support for multiple research projects, helping teams achieve new progress in cutting-edge studies. Recently, several achievements were published in top international journals such as Cell and Nature Plants, covering fields including insect-bacteria symbiotic systems, the neutrophil evolutionary cycle model, and motor degenerative diseases, reflecting the ongoing expansion and deepening of life science exploration. AtaGenix's technical services played a critical role in driving the continuous output of research results.
No. 1
Title: Bacterial tubular networks channel carbohydrates in insect endosymbiosis
Journal: Cell
Impact Factor: 42.5
Author Affiliation: Institut National des Sciences Appliquées de Lyon
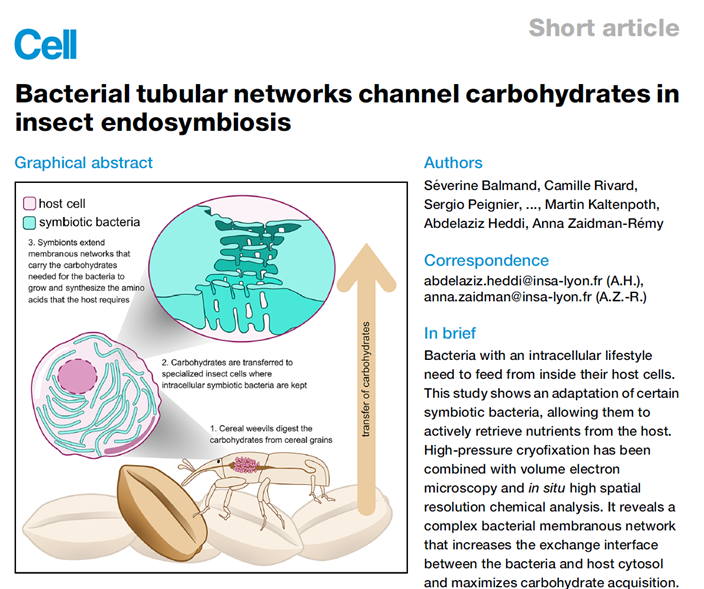
Symbiosis is widespread in nature, particularly nutritional endosymbiosis, where host cells (such as bacteriocytes in insects) harbor endosymbiotic bacteria, forming a "metabolic factory" that supplements the host's nutritional needs under restrictive diets through extensive metabolic exchange. Using the grain weevil and its endosymbiotic bacteria as a model, these bacteria provide the host with vitamins, amino acids, and other nutrients, especially producing phenylalanine and tyrosine from carbohydrate metabolism for host cuticle synthesis. Previous genomic and transcriptomic studies showed that endosymbiont genomes have few transporter genes, while host bacteriocytes are rich in carbohydrates, yet the exchange mechanism remained unclear. Through high-pressure freezing (HPF) combined with volume electron microscopy and in situ high-resolution chemical analysis, researchers revealed that endosymbiotic bacteria produce complex membrane tubular networks (tubenets) with diameters of about 20 nm. These structures connect bacteria and greatly expand the exchange interface with the host cytoplasm. Analysis showed that tubenets and host vesicles are rich in carbohydrates, indicating their role in efficiently acquiring sugar substrates provided by the host to support bacterial growth and nutrient synthesis. The study concludes that this bacterial strategy converges with multicellular organisms to enhance nutrient absorption; in the genus Sitophilus, the emergence of tubenets is linked to bacterial load and host ecological adaptation, emphasizing active participation of endosymbionts in metabolic exchange. This work highlights bacterial adaptation mechanisms in intracellular life and offers new perspectives on nutrient transfer in parasitic and mutualistic relationships.
AtaGenix customized a rabbit polyclonal anti-Lpp antibody for this study, used in immunogold labeling experiments to confirm that tubenets extend from the bacterial outer membrane rather than originating from the host. Combined with anti-LPS and anti-OmpC labeling, this formed "triple evidence," establishing tubenets as nutrient channels actively constructed by bacteria, directly supporting their function in efficient host carbohydrate uptake.
No. 2
Title: Conserved adaptor orchestrates co-secretion of synergistic type VI effectors in gut Bacteroidota
Journal: Cell Host & Microbe
Impact Factor: 18.7
Author Affiliation: Shandong University
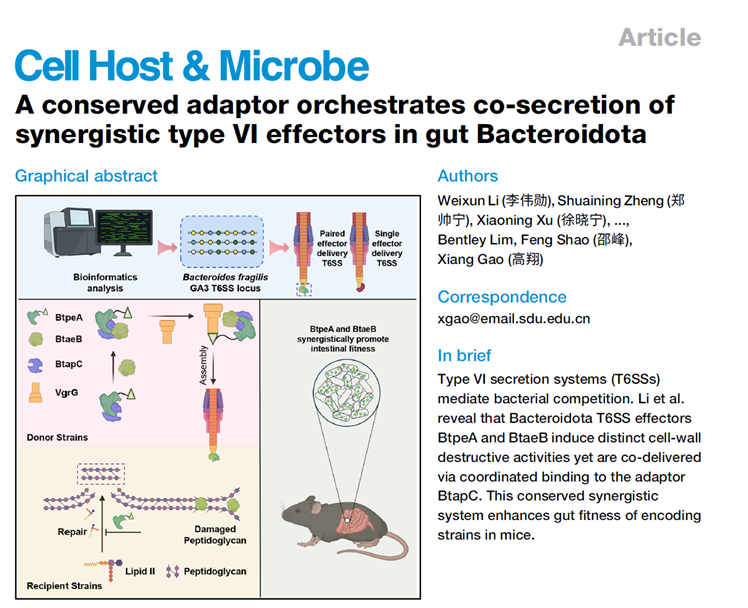
Gut Bacteroidota bacteria are key components of the human gut microbiota. They inject toxic effectors into neighboring bacteria via the type VI secretion system (T6SS) to gain a competitive edge in interspecies rivalry, thereby shaping gut microbial structure and influencing host health—this mechanism is a core focus of current gut microbiome research. This study delves into the secretion and functional mechanisms of Bacteroides T6SS effectors, identifying two complementary effectors in Bacteroides fragilis: BtpeA (a ColM-like phosphatase) and BtaeB (a peptidoglycan amidase). These do not act independently but form a stable complex with the conserved adaptor protein BtapC, enabling synergistic secretion via T6SS. Further validation confirmed that this "adaptor-mediated co-secretion of effectors" mechanism is widely conserved across gut Bacteroidota and significantly enhances bacterial colonization fitness in mouse intestines, providing a novel molecular foundation for understanding gut microbial interactions and developing microbiota modulation strategies.
AtaGenix customized four rabbit polyclonal antibodies targeting BtpeA, BtaeB, BtapC, and Hcp1 for this study. Prepared through antigen affinity purification and other processes, these antibodies exhibit high specificity and affinity for precise target protein recognition. Using them, researchers confirmed that BtpeA and BtaeB secretion depends on BtapC and correlates with T6SS activity (indicated by Hcp1); they also captured and identified the VgrG-BtpeA-BtaeB-BtapC quaternary complex, validating the molecular mechanism of adaptor BtapC-mediated effector assembly.
No. 3
Title: Pancentromere landscape and dynamic evolution in Brassica plants
Journal: Nature Plants
Impact Factor: 13.6
Author Affiliation: Peking University

Centromeres are critical structures for chromosome segregation with highly conserved functions, yet their sequences and proteins (such as CENH3) evolve rapidly across species. However, centromeres are rich in repetitive sequences, making assembly challenging and limiting understanding of their landscape. Brassica plants (e.g., Chinese cabbage, mustard, rapeseed) are closely related to Arabidopsis, exhibit diverse morphotypes, and include allopolyploids formed by hybridization, making them ideal for studying intra- and interspecies centromere evolution. By generating telomere-to-telomere (T2T) complete genome assemblies for seven morphotypes of B. rapa (AA genome), along with near-T2T assemblies for B. juncea (AABB) and B. napus (AACC), the researchers constructed a pan-centromere. Combined with CENH3 ChIP-seq, repeat annotation, and phylogenetic analysis, they dissected centromere structure, diversity, and evolution. Key findings: Brassica centromeres are widely invaded by retrotransposons with high size and structural diversity; A and C genome centromeres are dominated by 176 bp satellites but with different patterns, C genome satellites showing layered expansion; the B genome lacks satellites and is instead enriched in Copia elements, possibly due to CENH3 mutations causing satellite degradation. The study proposes a centromere evolutionary cycle model, reconstructing key events (e.g., satellite homogenization and transposon invasion), offering new insights into plant centromere evolution and guiding synthetic chromosome design in crops.
AtaGenix customized a key CENH3 antibody for this study, used in ChIP-seq experiments to immunoprecipitate chromatin from young leaves, identifying centromere positions, core regions, and CENH3 enrichment patterns. This supported the epigenetic definition of centromeres, analysis of satellite and transposon composition, and intra-/interspecies diversity comparisons, serving as core technical support for validating centromere function and evolutionary mechanisms.
No. 4
Title: Lysosomal damage is a therapeutic target in Duchenne muscular dystrophy
Journal: Science Advances
Impact Factor: 12.5
Author Affiliation: Université Paris Cité
DMD is an X-linked recessive muscular degenerative disease caused by the absence of dystrophin protein encoded by the DMD gene, leading to muscle fiber instability, calcium homeostasis disruption, oxidative stress, and fibroadipogenic replacement in a pathological cascade. Current gene therapies deliver micro-dystrophin (μDys) via adeno-associated virus (AAV); though FDA-approved, clinical efficacy is limited and fails to fully reverse all pathologies. The authors hypothesized that dystrophin deficiency-induced metabolic disorders may involve lysosomal dysfunction, which has been overlooked in muscular dystrophies. By detecting lysosomal membrane permeabilization (LMP) marker Gal-3 (LGALS3) in DMD patients and animal models, they found increased lysosomal numbers, enlarged morphology, reduced acidification, and declined proteolytic activity, alongside activated endolysosomal damage responses and autophagy impairment. High-cholesterol diets exacerbated LMP, while μDys gene therapy could not fully correct it; however, full-length γ-sarcoglycan gene therapy reversed it in another dystrophy (LGMDR5) model, suggesting LMP is more persistent in DMD. Using FDA-approved lysosomal protector trehalose combined with suboptimal-dose AAV-μDys in Dmdmdx mice significantly improved motor function, muscle histopathology, and transcriptome profiles. The study concludes that lysosomal damage is a key pathological mechanism in DMD; targeting it alongside gene therapy enhances efficacy, offering a novel combination treatment strategy.
AtaGenix developed a mouse anti-51r20 monoclonal antibody for this study, used in Meso Scale Discovery (MSD) ELISA assays. It served as a serum biomarker for assessing muscle damage in DMD mouse models, complementing lysosomal markers like Gal-3 to confirm correlations between lysosomal damage and myopathology, supporting improved overall muscle function with combination therapy—though not the core LMP detection method.
No. 5
Title: Improvement of photosynthetic efficiency and yield of upland cotton by SiFBA4 gene of Saussurea involucrata
Journal: Industrial Crops and Products
Impact Factor: 6.2
Author Affiliation: Shihezi University

Upland cotton (Gossypium hirsutum) is a major global natural fiber crop; improving its yield and quality is a breeding priority, with photosynthesis contributing ~95% to yield—enhancing photosynthetic efficiency is thus a key strategy. The Calvin cycle enzyme fructose-1,6-bisphosphate aldolase (FBA) plays roles in photosynthesis, sugar metabolism, and stress responses. Prior studies showed FBA influences plant growth, CO2 fixation, and yield, but comprehensive mechanisms remain limited. The Saussurea involucrata SiFBA4 gene previously enhanced cold tolerance in tomato and growth in tobacco; when transferred to cotton, it accelerated growth without significant cold tolerance differences. This study further evaluated its effects on photosynthesis, agronomic traits, yield, and fiber, using transcriptomics and metabolomics to elucidate mechanisms. Results showed transgenic lines with 17.47% higher net photosynthetic rate, 19.22% increased seed cotton yield, more fruit branches and bolls, but reduced fiber length and strength; metabolomics revealed decreased leaf sugars and increased root sugars; transcriptomics showed upregulation of Calvin cycle enzymes, chloroplast electron transport chain components (PSI, Cytb6/f, PSII), chlorophyll synthesis, and sugar transport genes. SiFBA4 boosts photosynthetic efficiency by enhancing Calvin cycle enzyme activity and light reaction gene expression, and optimizes photosynthate allocation via sugar transport gene upregulation, thereby increasing yield. This work provides genetic resources for cotton breeding and lays a foundation for designing crop carbon assimilation and allocation networks.
AtaGenix customized a rabbit monoclonal antibody against SiFBA4 for this study, used to detect SiFBA4 protein expression in proteins extracted from 7-day-old seedlings. It was a key reagent for verifying SiFBA4 protein expression in transgenic cotton, supporting molecular confirmation of positive transformants and providing a foundation for subsequent photosynthetic physiology, agronomic phenotyping, transcriptomic, and metabolomic analyses—ensuring transgene reliability and directly underpinning the core conclusion that "SiFBA4 enhances photosynthetic efficiency and yield."
No. 6
Title: Tudor-based proteomic strategy pan-specifically enriches and identifies protein arginine methylation
Journal: EMBO
Impact Factor: 6.2
Author Affiliation: School of Pharmaceutical Sciences, Sun Yat-sen University

Protein arginine methylation is a key post-translational modification in eukaryotes, regulating transcription, DNA damage repair, and other processes. Existing enrichment methods suffer from sequence bias or operational complexity, hindering comprehensive site capture. This study developed a molecular affinity strategy based on the SMN protein Tudor domain, pan-specific for mono- and di-methylated arginine, enriching methylated peptides from RGG/RG-rich regions and non-RG motifs with low sample requirements and simple operation. Using this approach, three dimethylation sites (R97, R99, R103) were identified in the RNA-binding domain of eukaryotic translation initiation factor eIF3D, with R99 being asymmetric dimethylation (aDMA) catalyzed by PRMT1. Functional validation showed that eIF3D R99 aDMA regulates cap-dependent translation initiation, positively modulating both classic eIF4E-dependent and c-JUN 5'UTR-mediated alternative pathways, revealing a novel role for arginine methylation in protein translation.
AtaGenix customized a rat polyclonal antibody specific for asymmetric dimethylation (aDMA) at eIF3D R99 (anti-R99me2a) for this study. Prepared using the modified peptide Cys-RNRMRme2aFAQRNL as immunogen and affinity-purified, it specifically recognizes aDMA at eIF3D R99 without binding unmodified peptides—a critical tool for validating this site's methylation status and laying the foundation for subsequent functional studies.
No. 7
Title: Stage-specific antiviral roles of IFIH1, IRF7, and MAVS in the interferon-mediated response to DHAV-3 infection
Journal: Poultry Science
Impact Factor: 4.2
Author Affiliation: Institute of Animal Sciences, Chinese Academy of Agricultural Sciences
Duck viral hepatitis (DVH) is a highly contagious disease; DHAV-3 (duck hepatitis A virus type 3) has become the dominant pathogen in East Asia in recent years, with unclear immune evasion mechanisms. The innate immune system, as the first line of defense, rapidly and nonspecifically recognizes pathogens while exhibiting specificity and memory features, yet dynamic innate immune studies in Peking ducks infected with DHAV-3 remain lacking. This study's core aim is to elucidate the regulatory network of innate immunity in Peking ducks post-DHAV-3 infection and the roles of key molecules, providing a theoretical basis for dissecting viral immune evasion and disease-resistant breeding. Focusing on innate immune molecules and their dynamic links with IFN-I (especially IFN-β), the team used silencing or overexpression to explore their effects on viral replication across infection stages. Conclusions reveal a time-dependent regulatory network in DHAV-3 interactions with the Peking duck innate immune system, with core molecules balancing host defense and viral exploitation through stage-specific functions—filling a gap in dynamic innate immunity research for DHAV-3 in Peking ducks and offering new insights for understanding viral immune mechanisms and resistance breeding.
AtaGenix customized rabbit monoclonal anti-IFN-α and anti-IFN-β antibodies for this study, used to detect IFN-α and IFN-β protein levels in primary duck hepatocytes post-DHAV-3 infection. These complemented qPCR gene-level results, confirming protein dynamic changes across infection stages and directly supporting key conclusions like "IFN-I significant upregulation with antiviral effects" and "IFIH1/MAVS/IRF7 regulation of IFN protein expression"—a vital technical link bridging transcription to protein-level validation of innate immune responses.
As of October 2025, over 500 publications have cited AtaGenix's one-stop protein and antibody development services. Looking ahead, AtaGenix will continue fully supporting scientific research, driving technological innovation and breakthroughs.
More Technical Services & Collaboration Inquiries
Feel free to contact: info@atagenix.com or call 027-87001869 for a tailored service plan.
Contact Us
+86-27-87001869
info@atagenix.com
Building C, R & D Building, No. 666, Shendun 4th Road, Donghu New Technology Development Zone, Wuhan

